-
What is this for?
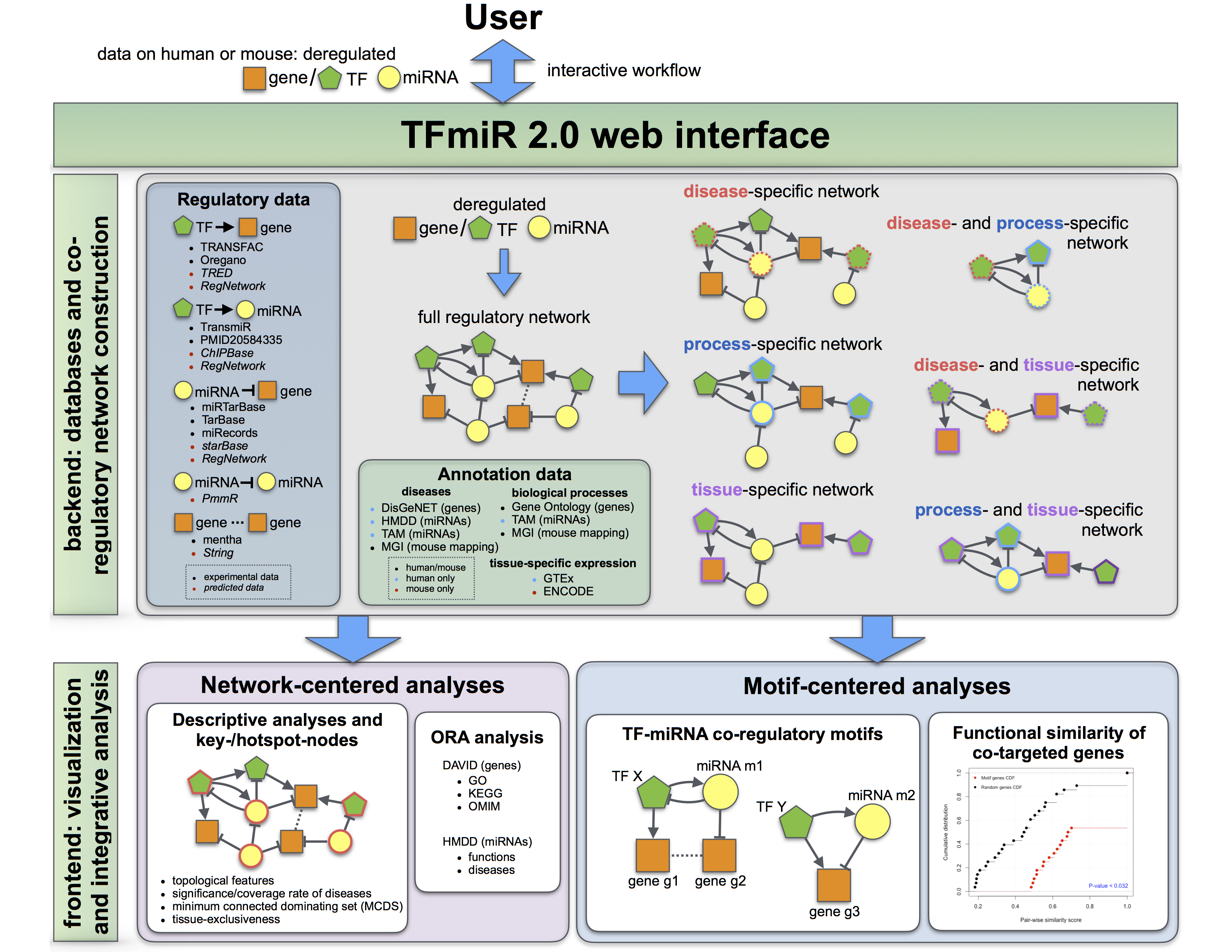
- For a list of up- and downregulated (1) mRNAs and (2) miRNAs from a differential analysis, TFmiR builds a co-regulatory network based on parameters set (3).
- Database retrieval of predicted and experimentally validated interactions, based on confidence level
- Generate complete, disease-, tissue-, process-, disease & process-, tissue & process- and disease & tissue-specific co-regulatory networks
-
What does the input look like?The input consists of
- a list of mRNA identifiers with an indicator for their up- or downregulation (-1/1)
- (optional) a list of miRNA identifiers with an indicator for their up- or downregulation (-1/1)
- parameters that determine the enrichment p-value (mRNA/miRNA), p-value treshold for the overrepresentation analysis, the associated disease, the associated process, the associated tissue, the confidence level (experimentally validated and/or predicted interactions), PPI/GGI threshold and randomization method for detection of motifs.
-
How exactly has a file to look like?An input file (for mRNA, miRNA analogous) looks like:
AATK -1 ABCB8 1 ABCG4 1 ABHD10 1 ABLIM1 -1 ABT1 1 etc...
However, you can try the dataset we used for breast neoplasm:
mirna.bc.txt
mrna.bc.txt
Alternatively, you can just click the "Load example data" at the start screen to load the dataset automatically. -
Which databases are incorporated into TFmiR2 for mouse?These databases are used:
(P) means predicted interactions and (E) means experimentally validated interactions.Interaction Databases (P/E) * Genes miRNAs Regulatory links Version /frozen date TF → gene TRRUST (E) [2] 2456 -- 6490 V2 OregAnno (E) [3] 22209 -- 54376 Nov 2016 ITFB (P) [15] 5118 -- 34679 -- TF → miRNA TransmiR (E) [5] 147 230 748 V2.0, Oct 2018 ChipBase (P) [6] 63 623 5764 -- TransmiR (E) [5] 534 1186 449894 V2.0, Oct 2018 miRNA → gene miRTarBase (E) [8] 846 292 1526 -- TarBase (E) [8] 83 44 122 -- miRecords (E) [9] 274 146 384 April 2013 starBase (P) [10] 1 84 98 -- gene → gene mentha(E) [13] 14328 -- 7325 June 2019 STRING(P) [14] 21291 -- 11944806 v11.0 -
I am still stuck!Contact us for help:
m.hamed@bioinformatik.uni-saarland.de
christian.spaniol@bioinformatik.uni-saarland.de
maryam.nazarieh@bioinformatik.uni-saarland.de
thorsten.will@bioinformatik.uni-saarland.de
Which databases are incorporated into TFmiR2 for human?
These databases are used:
| Interaction | Databases (P/E) * | Genes | miRNAs | Regulatory links | Version /frozen date |
| TF → gene | TRANSFAC (E) [1] | 1279 | -- | 2943 | V11.4 | TRRUST (E) [2] | 2862 | -- | 8427 | V2 |
| OregAnno (E) [3] | 23506 | -- | 215832 | Nov 2016 | |
| TRED (P) [4] | 3038 | -- | 6462 | 2007 | |
| TF → miRNA | TransmiR (E) [5] | 369 | 402 | 2678 | V2.0, Oct 2018 |
| PMID20584335 (E) [6] | 58 | 56 | 102 | Apr 2009 | |
| ChipBase (P) [7] | 119 | 1380 | 33087 | V1.1, Nov 2012 | |
| TransmiR (E) [5] | 600 | 1861 | 1203375 | V2.0, Oct 2018 | |
| miRNA → gene | miRTarBase (E) [8] | 2849 | 740 | 8489 | V7.0, Sep 2013 |
| TarBase (E) [9] | 422 | 79 | 492 | V7.0 | |
| miRecords (E) [10] | 1106 | 280 | 1716 | April 2013 | |
| starBase (P) [11] | 1 | 190 | 190 | V3.0, 2018 | |
| miRNA → miRNA | PmmR(P) [12] | -- | 312 | 3846 | Mar 2011 |
| gene → gene | mentha(E) [13] | 18861 | -- | 339559 | June 2019 |
| STRING(P) [14] | 19344 | -- | 11754228 | v11.0 |